%%HTML
<link rel="stylesheet" type="text/css" href="https://raw.githubusercontent.com/malkaguillot/Foundations-in-Data-Science-and-Machine-Learning/refs/heads/main/docs/utils/custom.css">
%%HTML
<link rel="stylesheet" type="text/css" href="../utils/custom.css">
Introduction: Decision Trees and Ensemble Learning¶
Decision Trees (DTs)¶
ML algorithms that progressively divide data sets into smaller data groups based on a descriptive feature, until they reach sets that are small enough to be described by some label.
One of the most powerful supervized machine learning method
Non-parametric
For classification and regression.
Oveview of this family of methods¶
Decision Trees¶
Decision Trees¶
A graphical representation of a segmentation of the predictor space into a number of simple regions
Decision trees learn a series of binary splits in the data based on hard thresholds.
- if yes, go right; if no, go left.
- Additional splits as you move through the tree.
What is a tree?¶
- Root node
- Splitting
- Branch
- Decision node (internal node)
- Leaf node (terminal node)
- Sub-tree
- Depth (level)
- Pruning
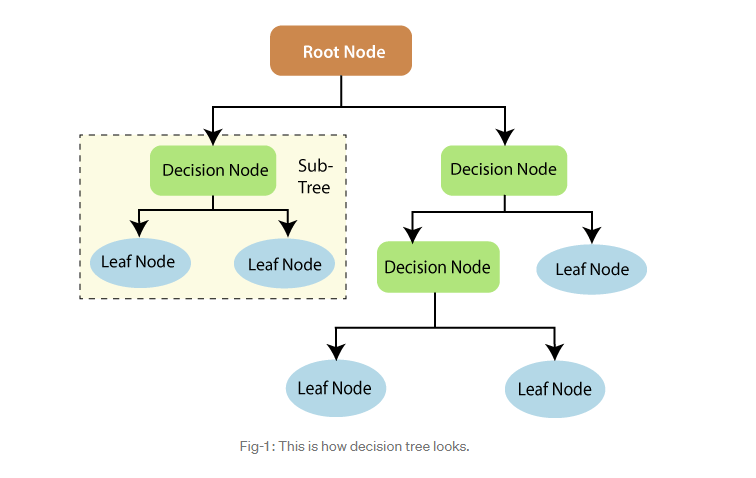
Decision Trees¶
- Classification trees:
- when predicting a discrete variable
- Example: predicting death after contracting Covid-19
- Regression trees:
- when predicting a continuous variable
- Example: predicting housing prices
hitters=pd.read_csv('../../data/Hitters.csv').dropna()
hitters.head()
| Unnamed: 0 | AtBat | Hits | HmRun | Runs | RBI | Walks | Years | CAtBat | CHits | ... | CRuns | CRBI | CWalks | League | Division | PutOuts | Assists | Errors | Salary | NewLeague | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | -Alan Ashby | 315 | 81 | 7 | 24 | 38 | 39 | 14 | 3449 | 835 | ... | 321 | 414 | 375 | N | W | 632 | 43 | 10 | 475.0 | N |
| 2 | -Alvin Davis | 479 | 130 | 18 | 66 | 72 | 76 | 3 | 1624 | 457 | ... | 224 | 266 | 263 | A | W | 880 | 82 | 14 | 480.0 | A |
| 3 | -Andre Dawson | 496 | 141 | 20 | 65 | 78 | 37 | 11 | 5628 | 1575 | ... | 828 | 838 | 354 | N | E | 200 | 11 | 3 | 500.0 | N |
| 4 | -Andres Galarraga | 321 | 87 | 10 | 39 | 42 | 30 | 2 | 396 | 101 | ... | 48 | 46 | 33 | N | E | 805 | 40 | 4 | 91.5 | N |
| 5 | -Alfredo Griffin | 594 | 169 | 4 | 74 | 51 | 35 | 11 | 4408 | 1133 | ... | 501 | 336 | 194 | A | W | 282 | 421 | 25 | 750.0 | A |
5 rows × 21 columns
Using 2 explanatory variable: Years & Hits¶
In particular, we are interested in looking how the number of Hits and the Years of experience predict the Salary.
# Get Features
features = ['Years', 'Hits']
X = hitters[features].values
y = np.log(hitters.Salary.values)
Salary vs. Log(Salary)¶
We are actually going to use log(salary) since it has a more gaussian distribution.
make_figure_hist()
make_figure_hits_year()
from sklearn.tree import DecisionTreeRegressor
from sklearn.tree._export import plot_tree
Regression tree with 3 leaves ie. 2 nodes
# Fit regression tree
tree = DecisionTreeRegressor(max_leaf_nodes=3)
tree.fit(X, y)
DecisionTreeRegressor(max_leaf_nodes=3)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeRegressor(max_leaf_nodes=3)
Visualize the tree¶
fig, ax = plt.subplots(1,1)
ax.set_title('Figure Decision tree');
# Plot tree
plot_tree(tree, filled=True, feature_names=features, fontsize=12, ax=ax);
A Visual Representation of the Data¶
make_figure_hits_year2()
Challenges with Splits¶
Using splits in general introduces three key questions:
- Where should the split be?
- How many splits are optimal?
- How to make predictions within each node?
These queries fall within the scope of the decision tree framework.
Building a Tree¶
Classification and regression trees (CART)¶
There are two main steps in the construction of a tree:
- We divide the predictor space—that is the set of possible values for $X_1, X_2, \cdots ,X_p$ into $J$ distinct and non-overlapping regions $R_1, R_2, \cdots, R_j$
- For every observation that falls into the region $R_j$, we make the same prediction, which is simply the mean of the response values for the training observations in $R_j$.
- For example, for a continuous $y$, $$\hat{y}_{j}=\frac{1}{N_{j}} \sum_{x_{i} \in R_{j}} y_{i}$$
The Splitting Process: How does it work?¶
Going over every possible partitions of the feature space is infeasible.
Instead, the CART algorithm follows a greedy approach.
Starting with all of the data, consider a splitting variable $j$ and split point $s$, and define the pair of half-planes $$R_{1}(j, s)=\left\{x | x_{j} \leq s\right\}, \qquad R_{2}(j, s)=\left\{x | x_{j}>s\right\}$$
Find the predictor $j^*$ and split $s^*$ that partitions the data into 2 regions $R_{1}(j^*,s^*)$ and $R_{2}(j^*,s^*)$ such that the overall sums of squares error are minimized: $$\mathrm{RSS}=\sum_{i \in R_{1}(j^*,s^*)}\left(y_{i}-\bar{y}_{1}\right)^{2}+\sum_{i \in R_{2}(j^*,s^*)}\left(y_{i}-\bar{y}_{2}\right)^{2}$$ where $\bar{y}_{1}$ and $\bar{y}_{2}$ are the averages of the training set outcomes within each group.
The CART Algorithm¶
Begin with the root node, containing the complete sample. Then:
Identify the single $RSS$ minimizing split for this node.
Split this parent node into the left and right node.
Apply steps 1 and 2 to each child node.
Repeat until reaching a leaf node of a pre-defined minimum size (for instance, cease splitting when fewer than 10 observations remain in each leaf).
How large should we grow the tree?¶
Large tree - overfit. Small tree - high variance.
The tree's level of expressiveness is captured by its size (the number of terminal nodes).
Common practice: Build a large tree and prune the tree backwards using cost-complexity pruning.
Cost-complexity pruning¶
The cost complexity criterion associated with a tree $T$ is given by
$$\mathrm{RSS}_{cp}(T)=\mathrm{RSS}(T)+cp|T|$$ where
- $\mathrm{RSS}$ is the sum of squared error for tree $T$.
- $|T|$ is the number of terminal nodes in tree $T$.
- $cp$ is the complexity parameter
- The complexity parameter is unit free and ranges from 0 to 1:
- When $cp = 0$, we have a fully saturated tree.
- When $cp = 1$, there are no splits, i.e, we predict the unconditional mean.
Hence, for CART, the penalty is a function of the number of terminal nodes.
NOTE: $cp$ and $|T|$ are analogous to $\lambda$ and $\lVert\boldsymbol{\beta}\rVert_1$ in the lasso.
Regularization Hyperparameters¶
- No parameter: nonparametric model
- To avoid overfitting control by the
max_depthhyperparametermax_depthmin_samples_split: the min number of samples a node must have before it can be splitmin_samples_leaf: the min number of samples a leaf node must have
# Compute tree
overfit_tree = DecisionTreeRegressor(max_leaf_nodes=5).fit(X, y)
# Plot tree
fig, ax = plt.subplots(1,1)
plot_tree(overfit_tree, filled=True, feature_names=features, fontsize=12, ax=ax);
Avoid overfitting using a minimum number of observations per leaf¶
# Compute tree
no_overfit_tree = DecisionTreeRegressor(max_leaf_nodes=5, min_samples_leaf=10).fit(X, y)
# Plot tree
fig, ax = plt.subplots(1,1)
plot_tree(no_overfit_tree, filled=True, feature_names=features, fontsize=14, ax=ax);
Choosing optimal tree length using cross-validation¶
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import train_test_split, KFold, cross_val_predict, cross_val_score
features = ['Years', 'Hits', 'RBI', 'PutOuts', 'Walks', 'Runs', 'AtBat', 'HmRun']
hitters=pd.read_csv('../../data/Hitters.csv').dropna()
X = hitters[features]
y =hitters['Salary']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
# Init
params = range(2,11)
reg_scores = np.zeros((len(params),3))
best_score = 10**6
# Loop over all parameters
for i,k in enumerate(params):
# Model
tree = DecisionTreeRegressor(max_leaf_nodes=k)
# Loop over splits
tree.fit(X_train, y_train)
reg_scores[i,0] = mean_squared_error(tree.predict(X_train), y_train)
reg_scores[i,1] = mean_squared_error(tree.predict(X_test), y_test)
# Get CV score
kf6 = KFold(n_splits=6)
reg_scores[i,2] = -cross_val_score(tree, X_train, y_train, cv=kf6, scoring='neg_mean_squared_error').mean()
# Save best model
if reg_scores[i,2]<best_score:
best_model = tree
best_score = reg_scores[i,2]
make_figure_optimal_depth()
The optimal tree depth is 4.
Classification¶
A classification tree is very similar to a regression tree, except that it is used to predict a qualitative response rather than a quantitative one.
For a classification tree, we predict that each observation belongs to the most commonly occurring class of training observations in the region to which it belongs.
Building a Classification Tree¶
- Similar to the task of growing a regression tree.
- However, RSS cannot be used as a criterion for making the binary splits.
- We define $\hat p_{mk}$ as the proportion of training observations in the $m^{th}$ region that are from the $k^{th}$ class.
Possible loss functions to decide the splits are:¶
Classification error rate $$ E = 1 - \max _{k}\left(\hat{p}_{m k}\right) $$
Gini index $$ G=\sum_{k=1}^{K} \hat{p}_{m k}\left(1-\hat{p}_{m k}\right) $$
Entropy $$ D=-\sum_{k=1}^{K} \hat{p}_{m k} \log \hat{p}_{m k} $$
Cons¶
- Poor predictive performance (typically)
- High risk of overfit
- Too many branches lower the variance but can increase the bias
- trees can be very non-robust.
- In other words, a small change in the data can cause a large change in the final estimated tree.
- trees can be very non-robust.
Extension: bagging, random forests, and boosting.¶
These methods grow multiple trees which are then combined to yield a single consensus prediction.
Combining a large number of trees $\rightarrow$
- dramatic improvements in prediction accuracy,
- at the expense of some loss interpretation.
XGBoost Ingredient 2 -- Bagging (Bootstrapping)¶
Bagging¶
The main problem of decision trees is that they suffer from high variance.
Bootstrap aggregation, or bagging, is a general-purpose procedure for reducing the variance of a statistical learning method.
- Averaging reduce the variance
Combines prediction from multiple decision trees through a majority votting mechanism
Out-of-Bag Error Estimation¶
Very straightforward way to estimate the test error of a bagged model, without the need to perform cross-validation or the validation set approach.
- Bagging: trees are repeatedly fit to bootstrapped subsets of the observations.
- On average, each bagged tree makes use of around two-thirds of the observations.
- The remaining one-third of the observations not used to fit a given bagged tree are referred to as the out-of-bag (OOB) observations.
- We can predict the response for the ith observation using each of the trees in which that observation was OOB.
Example in the case of a classification¶
We are now going to compute the Gini index for the Heart dataset using different numbers of trees.
We use the Heart dataset on individual heart failures. We will try to use individual characteristics in order to predict heart deseases (HD). The varaible is binary: Yes, No.
# Load heart dataset
heart=pd.read_csv('../../data/Heart.csv').drop('Unnamed: 0', axis=1).dropna()
heart.head()
| Age | Sex | ChestPain | RestBP | Chol | Fbs | RestECG | MaxHR | ExAng | Oldpeak | Slope | Ca | Thal | AHD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 63 | 1 | typical | 145 | 233 | 1 | 2 | 150 | 0 | 2.3 | 3 | 0.0 | fixed | No |
| 1 | 67 | 1 | asymptomatic | 160 | 286 | 0 | 2 | 108 | 1 | 1.5 | 2 | 3.0 | normal | Yes |
| 2 | 67 | 1 | asymptomatic | 120 | 229 | 0 | 2 | 129 | 1 | 2.6 | 2 | 2.0 | reversable | Yes |
| 3 | 37 | 1 | nonanginal | 130 | 250 | 0 | 0 | 187 | 0 | 3.5 | 3 | 0.0 | normal | No |
| 4 | 41 | 0 | nontypical | 130 | 204 | 0 | 2 | 172 | 0 | 1.4 | 1 | 0.0 | normal | No |
Pre-processing step¶
# Fastorize variables
heart.ChestPain = pd.factorize(heart.ChestPain)[0]
heart.Thal = pd.factorize(heart.Thal)[0]
# Set features
features = [col for col in heart.columns if col!='AHD']
X2 = heart[features]
y2 = pd.factorize(heart.AHD)[0]
from sklearn.ensemble import BaggingClassifier
from sklearn.tree import DecisionTreeClassifier
# Init (takes a lot of time with J=30)
params = range(2,50)
bagging_scores = np.zeros((len(params),2))
J = 30;
# Loop over parameters
for i, k in enumerate(params):
print("Computing k=%1.0f" % k, end ="")
# Repeat J
temp_scores = np.zeros((J,2))
for j in range(J):
X2_train, X2_test, y2_train, y2_test = train_test_split(X2, y2, test_size=0.5, random_state=j)
bagging = BaggingClassifier(DecisionTreeClassifier(), max_samples=k, oob_score=True)
bagging.fit(X2_train,y2_train)
temp_scores[j,0] = bagging.score(X2_test, y2_test)
temp_scores[j,1] = bagging.oob_score_
# Average
bagging_scores[i,:] = np.mean(temp_scores, axis=0)
print("", end="\r")
Computing k=49
Plot of the Out-of-Bag error computed while generating the bagged estimator.¶
make_new_figure()
Variable Importance Measures¶
Great property of random forests
Importance of features by looking at how much the tree nodes that use that feature reduce impurity on average (across all trees in the forest).
The measure relies on
- regression: the total amount of the RSS that is decreased due to splits over a given predictor, averaged over all trees.
- classification: we can add up the total amount that the Gini index is decreased by splits over a given predictor, averaged over all trees.
A large value indicates an important predictor.
SkLearncomputes this score automatically for each feature after training,- results scaled to sum to 1
feature_importances_
# Compute feature importance
feature_importances = np.mean([tree.feature_importances_ for tree in bagging.estimators_], axis=0)
make_figure_feature_importance()
XGBoost Ingredient 3 -- Random Forest¶
Random Forest¶
= an ensemble of Decision Trees
Each tree gets its own sample of data.
At each tree split, a random sample of features is drawn, only those features are considered for splitting.
For each tree, error rate is computed using data outside its bootstrap sample.
Random Forest classifier¶
Let's split the data in 2 and compute test and estimated $R^2$, for both forest and trees.
from sklearn.ensemble import RandomForestClassifier
# Init (takes a lot of time with J=30)
params = range(2,50)
forest_scores = np.zeros((len(params),2))
J = 30
# Loop over parameters
for i, k in enumerate(params):
print("Computing k=%1.0f" % k, end ="")
# Repeat J
temp_scores = np.zeros((J,2))
for j in range(J):
X2_train, X2_test, y2_train, y2_test = train_test_split(X2, y2, test_size=0.5, random_state=j)
forest = RandomForestClassifier(n_estimators=k, oob_score=True, max_features="sqrt")
forest.fit(X2_train,y2_train)
temp_scores[j,0] = forest.score(X2_test, y2_test)
temp_scores[j,1] = forest.oob_score_
# Average
forest_scores[i,:] = np.mean(temp_scores, axis=0)
print("", end="\r")
Computing k=49
make_figure_bagging_vs_rf()
As for bagging, we can plot feature importance.
make_figure_feature_importance2()
XGBoost Ingredient 4 -- Boosting¶
Boosting¶
Any Ensemble method that can combine several weak learners into a strong learner
trees are grown sequentially: each tree is grown using information from previously grown trees.
- Boosting does not involve bootstrap sampling; instead each tree is fit on a modified version of the original data set.
Most used boosting methods:
- Adaptive Boosting :
AdaBoost - Gradient Boosting
- Adaptive Boosting :
XGBoost Ingredient 5 -- Gradient Boosting¶
Gradient boosting = an additive ensemble of trees¶
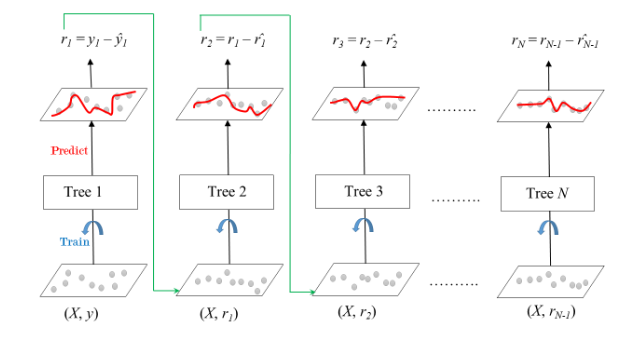
Adds additional layers of trees to fit the residuals of the first layers
XGBoost Ingredient 6 -- XGBoost¶
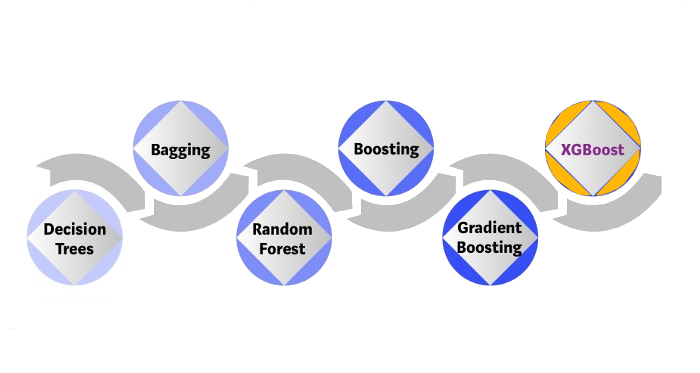
XGBoost =Extreme Gradient Boosting¶
$\rightarrow$ optimized implementation of Gradient Boosting
- easy to use
- actively developed
- efficient / parallelizable
- provides model explanations
- takes sparse matrices as input
import xgboost
xgb_reg=xgboost.XGBRegressor()
xgb_reg.fit(X_train, y_train)
y_pred=xgb_reg.predict(X_test)
Feature Importance¶
from xgboost import plot_importance
plot_importance(xgb_reg)
<Axes: title={'center': 'Feature importance'}, xlabel='Importance score', ylabel='Features'>
Random forests and boosted trees provide a metric of feature importance that summarizes how well each feature contributes to predictive accuracy.
